A Helper Virus-independent, Plasmid only-based Reverse Genetics System for Rotavirus
Ulrich Desselberger
Abstract
Compared to classical ‘forward’ genetics in which the changed phenotype of an organism is correlated with mutations in the genome, the availability of complete nucleotide sequences of many microbes and animals now allows the rational engineering of mutations in individual genes and identification of the correlated change in phenotype of the reconstituted organism (‘reverse genetics’). Reverse genetics enables precise assignment of phenotypic changes to genomic mutations. The goal to ‘rescue’ infectious rotavirus from a mutated genome has eluded success for a long time, and only recently a plasmid only-based, helper virus-free reverse genetics system for species A rotaviruses has been developed. Based on background information on rotavirus structure, classification, replication and genetic research, the new procedure is reviewed in detail, and its versatility and significance for future basic and translational research are discussed.
Introduction
Rotaviruses (RVs) are a major cause of acute gastroenteritis (AGE) in infants and young children worldwide and in many mammalian and avian species1,2. RV-associated disease led to the death of over 200,000 children of <5 years of age in 20133 and thus represents a major pediatric, public health and economic problem. Since 2006 two RV vaccines (Rotarix®, RotaTeq®) have been licensed, were included into national immunization programs in 82 countries (January 2016) and were found to be highly effective in preventing severe disease in high-income countries, less so in low- and middle-income countries4,5. Based on RV structure and classification, the viral replication cycle and viral genetics, the recent achievement of a plasmid only-based reverse genetics (RG) for RV enabling the recovery of infectious virus from cDNA which can be mutated precisely is briefly described and put into the larger context of RV research.
Rotavirus Structure and Classification
Rotaviruses are triple-layered particles containing 11 segments of double-stranded (ds) RNA as their genome. The RNA segments encode 6 structural proteins (VP1-VP4, VP6, VP7) and 5-6 non-structural proteins (NSP1-NSP5/6). The viral genome, the RNA-dependent RNA polymerase (VP1) and the capping enzyme (VP3) are surrounded by an inner protein layer (VP2), forming a core. This, in turn, is surrounded by an intermediate layer (VP6), to form a double-layered particle (DLP), and an outer layer, consisting of the VP7 protein and the spike-forming VP4, to become a triple-layered particle (TLP), the infectious virion1,2. (Figure 1)
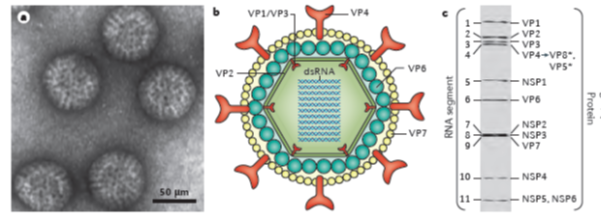
Figure 1: Rotavirus structure and gene-protein assignment (for simian RV SA11 strain)
a. Electron micrograph of triple-layered rotavirus particles.
b. Cross-sectional diagram of the rotavirus triple-layered particle: the viral genome, the enzymes VP1 and VP3 and the structural protein VP2 comprise the core, the middle layer is formed by VP6, and the outer layer by the proteins VP7 and VP4 (with the latter being proteolytically cleaved into VP5* and VP8*).
c. Electrophoretic separation profile of the 11 dsRNA segments, and gene-protein assignment for simian rotavirus SA11 strain (VP, viral protein; NSP, non-structural protein)
[From: Crawford, SE et al, Rotavirus infection. Nat Rev Dis Primers 2017; 3: 17083. [ref 49]
Rotaviruses constitute a genus of the Reoviridae family. According to the serological reactivity and genetic variability of VP6 at least 10 groups or species (A- I; J) have been differentiated6, and within species, various genotypes determined by the genes coding for VP7 (G types) and VP4 (P types) have been observed. More recently, a comprehensive classification system differentiating genotypes of all 11 RNA segments has been developed7,8. Thus, within species, A RVs (RVAs), at least 28 G types, 39 P types and between 14 and 24 genotypes for the remaining nine RNA segments were recognized in 20158.
Rotavirus Replication
The RV TLPs first attach to sialo-glycans or histo-blood group antigens (depending on strain) on the surface of susceptible host cells. Internalization of RV particles occurs by receptor-mediated endocytosis and is followed by removal of the outer layer in the endosome, resulting in the release of DLPs into the cytoplasm. The DLPs are transcription active and produce the full spectrum of 11 (+) single-stranded (ss) RNAs. Those either act as mRNAs from which viral proteins are translated, or become templates for the dsRNA replication of progeny virus. Two RV proteins, NSP2 and NSP5, are essential for the formation of cytoplasmic inclusion bodies termed ‘viroplasms,’ in which the viral RNA segments are assorted, replicated (to dsRNA) and packaged into new DLPs. Upon release from viroplasms, DLPs bind to NSP4, which is inserted in the endoplasmic reticulum (ER), serving as an intracellular receptor and mediating the transport of DLPs into the ER. NSP4 has pleiotropic properties, also acting as a viroporin to release Ca2+ from intracellular stores. In the ER DLPs acquire the outer capsid proteins VP4 and VP7 and become transiently enveloped, the mature infectious TLPs loose the envelope. The progeny virions are released from cells by lysis or, in polarized epithelial cells, by a non-classical vesicular transport mechanism1,2. (Figure 2)
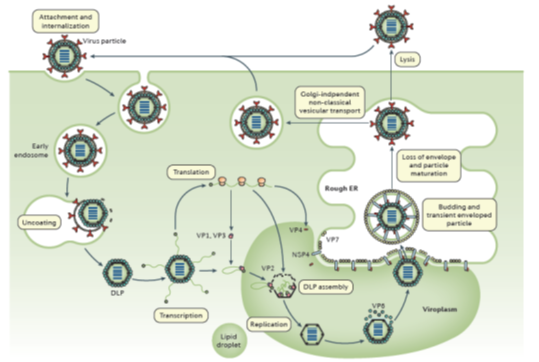
Figure 2: The rotavirus replication cycle
[From: Crawford, SE et al, Rotavirus infection. Nat Rev Dis Primers 2017; 3: 17083.]= ref 49
Rotavirus Genetics
The genetics of RVs are determined by their RNA segments and mutations therein. The ‘classical’ way to study virus mutants (occurring spontaneously or created by mutagenesis) consists of observing a change in phenotype in the mutant compared to the wild-type and correlating this with the mutant genotype (‘forward genetics’). Rotavirus mutants can be studied by complementation or reassortment analyses, e.g. for most of the temperature-sensitive (ts) RV mutants genetic mutations have been localized to individual RNA segments9. With the availability of the complete nucleotide sequences of most viral genomes, the strategy of genetic analysis of particular phenotypes has been completely transformed: Following rational engineering of particular mutations into individual viral genes at DNA or cDNA level, infectious viral particles of the genomic mutants can be ‘rescued’ and then phenotypically characterized (‘reverse genetics’).
Rotavirus Reverse genetics
Establishing reverse genetics systems for DNA viruses is comparatively easy since virtually all viral DNA genomes, which have been mutated in vitro, are infectious upon transfection. By contrast, the reverse genetics systems for RNA viruses are more complicated, requiring the following steps: in vitro reverse transcription of the genomic RNA into cDNA, in vitro mutagenesis of the cDNA, production of infectious progeny virus from the mutated cDNAs.
Helper virus-dependent reverse genetics system for rotavirus
For RVs, initial attempts to rescue a mutated or heterologous RNA segment succeeded by co- or superinfection with a helper virus10-12, e.g. a VP4-specific RVA [RVA is explained at the end of the section on RV classification] RNA, which was intracellularly transcribed from a transfected recombinant plasmid, was rescued by infection with a helper virus in the presence of VP4-specific antibodies strongly neutralizing the helper virus10. An alternative approach consisted of a dual selection system, in which a ts RVA mutant was used as helper virus at the non-permissive temperature; and additional selection pressure was created by the presence of a siRNA specifically directed against the helper virus gene (NSP2 in11), but not the heterologous NSP2 gene which was transcribed intracellularly from a transfected recombinant plasmid and intended to replace the cogent authentic gene of the helper virus11. This system also permitted the rescue of engineered partial viral RNA segment duplications (i.e., rearrangements) or heterologous cDNA sequences from the intracellularly transcribed recombinant RNA segment12.
A plasmid only-based reverse genetics system for rotavirus
Kanai et al.13 have now developed a plasmid only-based reverse genetics system for RVAs. Using an approach that had previously been successful with orthoreoviruse14, Kanai et al. 13 constructed plasmids, which each contained the cDNA of one of the 11 RV RNA segments (SA11 strain) inserted between a T7 RNA polymerase (T7Pol) promoter at the 5’end and the antigenomic hepatitis delta virus ribozyme at the 3’end. Upon co-transfection into BHK cells constitutively expressing T7Pol, authentic full-length viral ss(+)RNA transcripts were expected to be synthesized. While this procedure (or a modification there of which used ss(+)RNAs transcribed from cDNA clones in vitro) was successful in ‘rescuing’ infectious orthoreovirus14 or bluetongue virus15 progeny, it did not lead to the recovery of infectious RV13,16. Progress was reached based on previous findings that fusion-associated small transmembrane (FAST) proteins (encoded by many Aquareovirus and some Orthoreovirus species)17 increased the yield of heterologous mammalian orthoreovirus (MRV)13 and of RV13 in infected cells substantially. Furthermore, the overexpression of the vaccinia virus (VV) capping enzyme (CE) complex increased the translatability of orthoreovirus (+) ssRNAs and greatly improved mammalian orthoreovirus rescue in the established reverse genetics systems13,14. When the 11 RV cDNA-containing recombinant plasmids and three plasmids expressing a reovirus FAST protein and the two subunits of the VV CE were co-transfected into baby hamster kidney (BHK) cells constitutively expressing T7Pol (BHK-T7) and lysates of transfected cells were passaged in African green monkey kidney (MA104) cells, infectious RV particles were recovered13, which turned out to be genuine and not a contaminant13. The single step growth kinetics of the rescued RV was indistinguishable from that of the parent virus. (Figure 3)
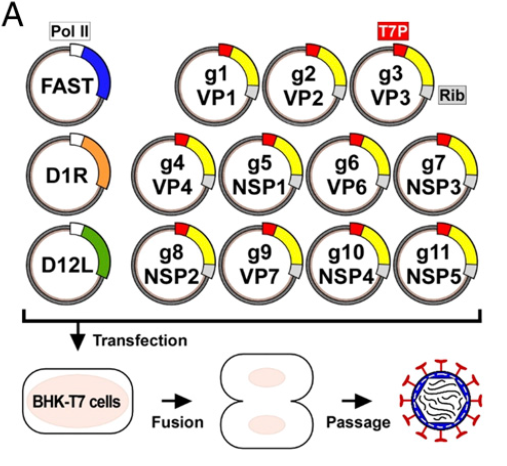
Figure 3: Development of a plasmid-based reverse genetics system for rotavirus
Strategy: RV cDNAs representing each of the 11 full-length rotavirus gene segments are flanked by the T7 polymerase promoter (T7P) and the antigenomic hepatitis delta virus ribozyme (Rib). BHK-T7 cells were transfected with the 11 rotavirus cDNAs and polymerase II promoter (Pol II)-driven expression plasmids encoding FAST and the vaccinia virus capping enzyme subunits (D1R and D12L). After 3-5 days of incubation, transfected cells were subjected to 3 cycles of freezing and thawing, and lysates were passaged on MA104 cells. A cytopathic effect (CPE) which developed after a few days, contained rescued infectious rotavirus.
(From: Kanai Y et al., Proc Natl Acad Sci USA 2017; 114(9): 2349-2354. = ref 13)
Versatility of the plasmid only-based reverse genetics system for rotavirus
This helper virus-free, entirely plasmid-based and fully tractable reverse genetics system permitted the recovery of RVs with genes reassorted as chosen and in combinations not encountered in nature13. The genetic background of this system is completely controlled, and unambiguous genotype - phenotype correlations can be achieved. Using the novel procedure Kanai et al.13 were able to explore the functions of the RV-encoded protein NSP1. Exploiting the split GFP system for fluorescent signalling18, one of the two GFP subunit genes was fused to the RV NSP1 gene (downstream of nucleotides encoding the C terminus of the NSP1 ORF), and the other one was expressed from a transfected plasmid; a viable recombinant RV was rescued, and both GFP subunits were functionally active by complementing each other in mutant RV-infected cells13. Furthermore, based on previous findings of the non-essential role of NSP1 for RV replication in vitro, an RV was constructed which expressed a foreign gene (encoding NanoLuc®-luciferase) fused to the NSP1 gene: the rescued RV recombinant supported stable expression of the transgenic nano-luciferase providing a bioluminescent signal, and this recombinant was used for dose-dependent screening of antivirals (here: ribavirin)13. Since its description in early 2017, the method is in the process of being established in some laboratories. So far, one publication using the system has appeared in which it was shown that the RV non-structural protein NSP6 is not essential for RV replication in cell culture19.
Reverse genetics systems of other RNA viruses
The work by Kanai et al.13 is the most recent addition to a large number of plasmid only-based reverse genetics systems of RNA viruses which were established for: poliovirus20, astrovirus21, coronavirus22, hepatitis C virus23, murine norovirus24; dimeric (+)ssRNA viruses: human immunodeficiency virus25; non-segmented and segmented (-)ssRNA viruses: rabies virus26, measles virus27, respiratory syncytial virus28, influenza A, B and C viruses29-32, Ebola virus33, bornavirus34; ambisense (+/-)ssRNA viruses: bunyavirus35; dsRNA viruses: infectious bursal disease virus36, orthoreovirus14, bluetongue virus15,37; and other viruses in each of these RNA virus groups.
Generally, helper virus-independent reverse genetics systems of RNA viruses have been substantially improved after their first description, e.g., by positioning the cDNAs of several or all RNA segments on one plasmid, thus reducing the number of plasmids to be co-transfected38-40, by staggered transfection41, or by the use of cell lines constitutively expressing T7 Pol38,42. The enormous possibilities of helper virus-free reverse genetics systems for different RNA viruses, but also some of the difficulties and challenges encountered in such work have been reviewed recently43.
Future perspectives
The work by Kanai et al.13 represents a key achievement in RV research and is very exciting. The novel helper virus-free reverse genetics system for RVs will enable the exploration of open questions of viral replication (e.g., genome assembly and packaging signals, and particle maturation) and of complex phenotypes (virulence, host range restriction, attenuation, a.o.). The procedure has recently been used to explore functions of the NSP6, encoded by RNA 11 from a second open reading frame: it suppresses the innate immune response by a so far unknown mechanism19. In the longer perspective, the RVA RG system has the potential of developing a ‘universal RV vaccine’, based on the recent discovery of highly cross-reactive and cross-neutralizing antibody producing B cells in the gut submucosa mediating heterotypic immunity44. It is expected that the availability of helper virus-free RG systems will move the field of vaccine design in a way similar to that of influenza viruses45-47 and of other RNA viruses48.
It can be expected that the plasmid only-based reverse genetics system for RVAs will substantially move the field forward in many laboratories as reverse genetics systems for other RNA viruses have done as soon as they were established.
References
- Estes MK, Greenberg HB. Rotaviruses. In: Knipe DM, Howley PM, et al., editors. Fields Virology, 6th ed. p. 1347–1401. Philadelphia: Wolters Kluwer Health/ Lippincott Williams & Wilkins; 2013.
- Desselberger U. Rotaviruses. Virus Res. 2014; 190: 75-96.
- Tate JE, Burton AH, Boschi-Pinto C et al. Global, regional, and national estimates of rotavirus mortality in children <5 years of age, 2000-2013.Clin Infect Dis. 2016; 62 Suppl 2: S96-S105.
- Jonesteller CL, Burnett E, Yen C, et al. Effectiveness of Rotavirus Vaccination: A systematic review of the first decade of global post-licensure data, 2006-2016. Clin Infect Dis. 2017 Apr 21. doi: 10.1093/cid/cix369. [Epub ahead of print]
- Burnett E, Jonesteller CL, Tate JE, et al. Global Impact of Rotavirus Vaccination on Childhood Hospitalizations and Mortality From Diarrhea. J Infect Dis. 2017 Jun 1 ;215(11): 1666-1672.
- Matthijnssens J, Otto PH, Ciarlet M, et al. VP6-sequence-based cutoff values as a criterion for rotavirus species demarcation. Arch Virol. 2012; 157(6):1177-1182.
- Matthijnssens J, Ciarlet M, Heiman E, et al. Full genome-based classification of rotaviruses reveals a common origin between human Wa-Like and porcine rotavirus strains and human DS-1-like and bovine rotavirus strains. J Virol. 2008; 82(7):3204-3219.
- https://rega.kuleuven.be/cev/viralmetagenomics/virus-classification/7th-RCWG-meeting
- Criglar J, Greenberg HB, Estes MK, et al. Reconciliation of rotavirus temperature-sensitive mutant collections and assignment of reassortment groups D, J, and K to genome segments. J Virol. 2011; 85(10): 5048-5060.
- Komoto S, Sasaki J, Taniguchi K. Reverse genetics system for introduction of site-specific mutations into the double-stranded RNA genome of infectious rotavirus. Proc Natl Acad Sci USA. 2006; 103(12): 4646-4651.
- Trask SD, Taraporewala ZF, Boehme KW, et al. Dual selection mechanisms drive efficient single-gene reverse genetics for rotavirus. Proc Natl Acad Sci USA. 2010; 107(43):18652-7.
- Navarro A, Trask SD, Patton JT. Generation of genetically stable recombinant rotaviruses containing novel genome rearrangements and heterologous sequences by reverse genetics. J Virol. 2013; 87(11):6211-6220.
- Kanai Y, Komoto S, Kawagishi T, et al. Entirely plasmid-based reverse genetics system for rotaviruses. Proc Natl Acad Sci USA. 2017; 114(9):2349-2354.
- Kobayashi T, Antar AA, Boehme KW, et al. A plasmid-based reverse genetics system for animal double-stranded RNA viruses. Cell Host Microbe. 2007; 1(2):147-157. Erratum in: Cell Host Microbe. 2(2):139 (2007).
- Boyce M, Celma CC, Roy P. Development of reverse genetics systems for bluetongue virus: recovery of infectious virus from synthetic RNA transcripts. J Virol. 2008; 82(17):8339-8348.
- Richards JE, Desselberger U, Lever AM. Experimental pathways towards developing a rotavirus reverse genetics system: synthetic full length rotavirus ssRNAs are neither infectious nor translated in permissive cells. PLoS One. 2013; 8(9):e74328.
- Ciechonska M, Duncan R. Reovirus FAST proteins: virus-encoded cellular fusogens. Trends Microbiol. 2014; 22(12):715-724.
- Cabantous S, Terwilliger TC, Waldo GS. Protein tagging and detection with engineered self-assembling fragments of green fluorescent protein. Nat Biotechnol. 2005; 23(1):102-107.
- Komoto S, Kanai Y, Fukuda S, et al. Reverse genetics system demonstrates that rotavirus nonstructural protein NSP6 is not essential for viral replication in cell culture. J Virol. 2017 Oct 13; 91(21). pii: e00695-17. doi: 10.1128/JVI.00695-17. Print 2017 Nov 1. PMID: 28794037.
- Racaniello VR, Baltimore D. Cloned poliovirus complementary DNA is infectious in mammalian cells. Science. 1981; 214(4523): 916-919.
- Geigenmüller U, Ginzton NH, Matsui SM. Construction of a genome-length cDNA clone for human astrovirus serotype 1 and synthesis of infectious RNA transcripts. J Virol. 1997; 71(2): 1713-1717.
- Almazán F, González JM, Pénzes Z, et al. Engineering the largest RNA virus genome as an infectious bacterial artificial chromosome. Proc Natl Acad Sci USA. 2000; 97(10):5516-5521.
- Wakita T, Pietschmann T, Kato T et al. Production of infectious hepatitis C virus in tissue culture from a cloned viral genome. Nat Med. 2005; 11(7): 791-796.
- Chaudhry Y, Skinner MA, Goodfellow IG. Recovery of genetically defined murine norovirus in tissue culture by using a fowlpox virus expressing T7 RNA polymerase. J Gen Virol. 2007; 88(Pt 8): 2091-2100.
- Sadaie MR, Rappaport J, Benter T, et al. Missense mutations in an infectious human immunodeficiency viral genome: functional mapping of tat and identification of the rev splice acceptor. Proc Natl Acad Sci USA. 1988; 85(23): 9224-9228.
- Schnell MJ, Mebatsion T, Conzelmann KK. Infectious rabies viruses from cloned cDNA. EMBO J. 1994; 13(18):4195-4203.
- Radecke F, Spielhofer P, Schneider H et al. Rescue of measles viruses from cloned DNA. EMBO J. 1995; 14(23):5773-5784.
- Collins PL, Hill MG, Camargo E et al. Production of infectious human respiratory syncytial virus from cloned cDNA confirms an essential role for the transcription elongation factor from the 5' proximal open reading frame of the M2 mRNA in gene expression and provides a capability for vaccine development. Proc Natl Acad Sci USA. 1995; 92(25):11563-11567.
- Neumann G, Watanabe T, Ito H, et al. Generation of influenza A viruses entirely from cloned cDNAs. Proc Natl Acad Sci USA. 1999; 96(16): 9345-9350.
- Fodor E, Devenish L, Engelhardt OG, et al. Rescue of influenza A virus from recombinant DNA. J Virol. 1999; 73(11): 9679-9682.
- Hoffmann E, Mahmood K, Yang CF, et al. Rescue of influenza B virus from eight plasmids. Proc Natl Acad Sci USA. 2002; 99(17):11411-11416.
- Crescenzo-Chaigne B, van der Werf S. Rescue of influenza C virus from recombinant DNA. J Virol. 2007; 81(20):11282-11289.
- Volchkov VE, Volchkova VA, Muhlberger E, et al. Recovery of infectious Ebola virus from complementary DNA: RNA editing of the GP gene and viral cytotoxicity. Science. 2001; 291(5510):1965-1969.
- Schneider U, Schwemmle M, Staeheli P. Genome trimming: a unique strategy for replication control employed by Borna disease virus. Proc Natl Acad Sci USA. 2005; 102(9):3441-3446.
- Bridgen A, Elliott RM. Rescue of a segmented negative-strand RNA virus entirely from cloned complementary DNAs. Proc Natl Acad Sci USA. 1996; 93(26): 15400-15404.
- Mundt E, Vakharia VN. Synthetic transcripts of double-stranded Birnavirus genome are infectious. Proc Natl Acad Sci USA. 1996; 93(20): 11131-11136.
- Pretorius JM, Huismans H, Theron J. Establishment of an entirely plasmid-based reverse genetics system for Bluetongue virus. Virology. 2015; 486:71-77.
- Kobayashi T, Ooms LS, Ikizler M, et al. An improved reverse genetics system for mammalian orthoreoviruses. Virology. 2010; 398(2): 194-200.
- Neumann G, Fujii K, Kino Y, et al. An improved reverse genetics system for influenza A virus generation and its implications for vaccine production. Proc Natl Acad Sci USA. 2005; 102(46): 16825-16829.
- Zhang X, Curtiss R 3rd. Efficient generation of influenza virus with a mouse RNA polymerase I-driven all-in-one plasmid. Virol J. 2015; 12:95.
- Matsuo E, Roy P. Bluetongue virus VP6 acts early in the replication cycle and can form the basis of chimeric virus formation. J Virol.83(17):8842-8848 (2009).
- Lowen AC, Noonan C, McLees A, et al. Efficient bunyavirus rescue from cloned cDNA. Virology. 2004; 330(2):493-500.
- Bridgen A, ed. Reverse genetics of RNA viruses. Applications and Perspectives. 408 pages. Wiley Blackwell, Hoboken NJ. 2012.
- Nair N, Feng N, Blum LK, et al. VP4- and VP7-specific antibodies mediate heterotypic immunity to rotavirus in humans. Sci Transl Med. 2017 Jun 21; 9(395). pii: eaam5434. doi 10.1126/ scitranslmed.aam5434.
- Nogales A, Martínez-Sobrido L. Reverse genetics approaches for the development of influenza vaccines. Int J Mol Sci. 2016 Dec 22; 18(1). pii: E20.
- Naito T, Mori K, Ushirogawa H, et al. Generation of a genetically stable high-fidelity influenza vaccine strain. J Virol. 2017 Feb 28; 91(6). pii: e01073-16.
- Stadlbauer D, Nachbagauer R, Meade P, et al. Universal influenza virus vaccines: what can we learn from the human immune response following exposure to H7 subtype viruses. Front Med. 2017 Nov 20. doi: 10.1007/s11684-017-0602-z. [Epub ahead of print]
- Stobart CC, Moore ML. RNA virus reverse genetics and vaccine design. Viruses. 2014 Jun 25; 6(7): 2531-50.
- Crawford SE, Ramani S, Tate JE, et al. Rotavirus infection. Nat Rev Dis Primers. 2017 Nov 9; 3: 17083.
